Note
Go to the end to download the full example code.
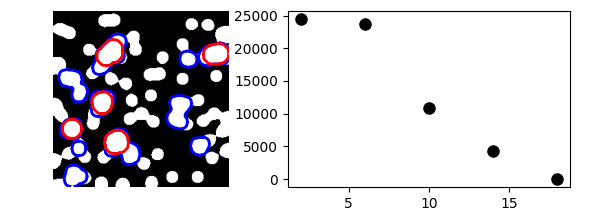
2.6.8.23. Granulometry¶
This example performs a simple granulometry analysis.
import numpy as np
import scipy as sp
import matplotlib.pyplot as plt
def disk_structure(n):
struct = np.zeros((2 * n + 1, 2 * n + 1))
x, y = np.indices((2 * n + 1, 2 * n + 1))
mask = (x - n) ** 2 + (y - n) ** 2 <= n**2
struct[mask] = 1
return struct.astype(bool)
def granulometry(data, sizes=None):
s = max(data.shape)
if sizes is None:
sizes = range(1, s / 2, 2)
granulo = [
sp.ndimage.binary_opening(data, structure=disk_structure(n)).sum()
for n in sizes
]
return granulo
rng = np.random.default_rng(27446968)
n = 10
l = 256
im = np.zeros((l, l))
points = l * rng.random((2, n**2))
im[(points[0]).astype(int), (points[1]).astype(int)] = 1
im = sp.ndimage.gaussian_filter(im, sigma=l / (4.0 * n))
mask = im > im.mean()
granulo = granulometry(mask, sizes=np.arange(2, 19, 4))
plt.figure(figsize=(6, 2.2))
plt.subplot(121)
plt.imshow(mask, cmap="gray")
opened = sp.ndimage.binary_opening(mask, structure=disk_structure(10))
opened_more = sp.ndimage.binary_opening(mask, structure=disk_structure(14))
plt.contour(opened, [0.5], colors="b", linewidths=2)
plt.contour(opened_more, [0.5], colors="r", linewidths=2)
plt.axis("off")
plt.subplot(122)
plt.plot(np.arange(2, 19, 4), granulo, "ok", ms=8)
plt.subplots_adjust(wspace=0.02, hspace=0.15, top=0.95, bottom=0.15, left=0, right=0.95)
plt.show()
Total running time of the script: (0 minutes 0.244 seconds)